# loading necessary libraries
library(here)
library(sf)
library(terra)
library(raster)
library(dplyr)
library(rpart)
library(rpart.plot)
library(tmap)Finding Suitable Locations for Marine Aquaculture
Marine aquaculture holds tremendous promise in addressing the global demand for sustainable protein sources, surpassing conventional land-based meat production. Recent research conducted by Gentry et al. has shed light on the vast potential of marine aquaculture worldwide, highlighting that all current wild-capture fisheries could be replaced by using less than 0.015% of the global ocean area for acquaculture (Gentry et al. 2017).
In this post, I embark on a journey to identify the most suitable locations along the west coast of the US for cultivating various marine life. I start by looking at habitat suitable for oysters, and end with a reproducible and reusable workflow workable for any species. My objective is to pinpoint various exclusive economic zones (EEZ)1 that have ideal conditions for marine aquaculture given each desired species ideal sea surface temperature and sea depth ranges.
A note on study limitations: According to Gentry et al., finding suitable marine aquaculture sites also depends on dissolved oxygen levels, and human-induced constraints such as protected areas, shipping lanes, and oil rigs. According to the study, the lethal limit of dissolved oxygen was not found to be a limiting factor for bivalves (mollusks such as oysters, clams, mussles, an scallops) and dissolved oxygen data is therefore left out of these analyses for simplicity. Human-induced constraints were also left out of this study but should be included for further analysis (Gentry et al. 2017).
Metadata
Sea Surface Temperature
I use average annual sea surface temperature (SST) from the years 2008 to 2012 to characterize the average sea surface temperature within the region. These data were originally collected from NOAA’s 5km Daily Global Satellite Sea Surface Temperature Anomaly v3.1.
Bathymetry
To characterize the depth of the ocean I use the General Bathymetric Chart of the Oceans (GEBCO).2
Exclusive Economic Zones
I use exclusive economic zonal boundaries from Marineregions.org.
The Code
Loading the Libraries
Loading and Cleaning Up the Data
Here I read in all the spatial data: west coast zone data, yearly sea surface temperature data, and approximate depth data. I then clean the data by (1) checking and aligning coordinate reference systems, map projections, spatial extent, and spatial resolutions (so that our data are correctly aligned in space) and (2) converting data types and units (for dataset compatibility and ease of future calculations).
You’ll see that I use the nearest neighbors approach to resample the depth data to match the resolution of the sea surface temperature data, which is of lower resolution. Downsampling like this is a common practice with geospatial datasets since the nearest neighbors approach preserves the original data by assigning the closest pixel in the higher resolution dataset (in this case the closest depth data point) to the newly created lower resolution pixel. I chose this method primarily due to its computational efficiency. Since depth data is continuous, for more accurate analysis something like bilinear or cubic interpolation may be better here.
# set working directory to here
#setwd(here())
# read in the shapefile of the maritime boundaries of the west coast region
wc_regions <- st_read(here("posts", "suitable-habitats", "data", "wc_regions_clean.shp"))Reading layer `wc_regions_clean' from data source
`/Users/victoriacutler/Documents/MEDS/victoriacutler.github.io/posts/suitable-habitats/data/wc_regions_clean.shp'
using driver `ESRI Shapefile'
Simple feature collection with 5 features and 5 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -129.1635 ymin: 30.542 xmax: -117.097 ymax: 49.00031
Geodetic CRS: WGS 84# read in the yearly sea surface temperature (SST) raster layers
average_annual_sst_2008 <- rast(here("posts", "suitable-habitats", "data", "average_annual_sst_2008.tif"))
average_annual_sst_2009 <- rast(here("posts", "suitable-habitats", "data", "average_annual_sst_2009.tif"))
average_annual_sst_2010 <- rast(here("posts", "suitable-habitats", "data", "average_annual_sst_2010.tif"))
average_annual_sst_2011 <- rast(here("posts", "suitable-habitats", "data", "average_annual_sst_2011.tif"))
average_annual_sst_2012 <- rast(here("posts", "suitable-habitats", "data", "average_annual_sst_2012.tif"))
# combine rasters into a stack
avg_annual_sst_stack <- stack(c(average_annual_sst_2008, average_annual_sst_2009, average_annual_sst_2010, average_annual_sst_2011, average_annual_sst_2012))
# read in the bathymetry (aka depth) raster
depth <- rast(here("posts", "suitable-habitats", "data", "depth.tif"))
# check the coordinate reference systems (CRSs)
cat(crs(wc_regions))GEOGCRS["WGS 84",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
CS[ellipsoidal,2],
AXIS["geodetic latitude (Lat)",north,
ORDER[1],
ANGLEUNIT["degree",0.0174532925199433]],
AXIS["geodetic longitude (Lon)",east,
ORDER[2],
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4326]]cat(as.character(crs(avg_annual_sst_stack)))+proj=longlat +ellps=WGS84 +no_defscat(crs(depth))GEOGCRS["WGS 84",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
CS[ellipsoidal,2],
AXIS["geodetic latitude (Lat)",north,
ORDER[1],
ANGLEUNIT["degree",0.0174532925199433]],
AXIS["geodetic longitude (Lon)",east,
ORDER[2],
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4326]] # converting the STT stack back to a SpatRaster to use terra functions, like to change the crs
avg_annual_sst_stack <- rast(avg_annual_sst_stack)
# converting the STT stack SpatRast to the matching crs
crs(avg_annual_sst_stack) = "EPSG:4326"
# find the mean SST then convert to Celcius
average_sst_degC <- mean(avg_annual_sst_stack, na.rm = TRUE) - 273.15
# crop the depth raster to match the extent of the SST raster
depth_newextent <- crop(depth, average_sst_degC) # you can see that the extents are now the same
# resample the depth raster such that the resolution is the same as the resolution in the SST raster
depth_newres <- resample(x = depth_newextent, y = average_sst_degC, method = "near")
# checking that the dimensions, resolutions, extent, and crs match
average_sst_degCclass : SpatRaster
dimensions : 480, 408, 1 (nrow, ncol, nlyr)
resolution : 0.04166185, 0.04165702 (x, y)
extent : -131.9848, -114.9867, 29.99305, 49.98842 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source(s) : memory
name : mean
min value : 4.980
max value : 32.895 depth_newresclass : SpatRaster
dimensions : 480, 408, 1 (nrow, ncol, nlyr)
resolution : 0.04166185, 0.04165702 (x, y)
extent : -131.9848, -114.9867, 29.99305, 49.98842 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source(s) : memory
name : depth
min value : -5468
max value : 4218 # doing this by checking to see that the two rasters can be stacked
stack_test <- rast(c(average_sst_degC, depth_newres))
stack_test # stackableclass : SpatRaster
dimensions : 480, 408, 2 (nrow, ncol, nlyr)
resolution : 0.04166185, 0.04165702 (x, y)
extent : -131.9848, -114.9867, 29.99305, 49.98842 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326) Using Reclassification Matrices to Assess Oyster Ecosystem Suitability
In this process, I conduct a reclassification of both the sea surface temperature raster and depth raster. Areas with suitable depth or temperature for oyster aquaculture are assigned a value of ‘1’, while unsuitable areas are marked as ‘NA’. By subsequently employing the lapply() function, I perform a multiplication operation on the reclassified rasters. The outcome is a final raster presenting a clear distinction between ‘1s’ indicating suitable habitat and ‘NAs’ representing areas unsuitable for oyster cultivation, providing a comprehensive depiction of overall suitability.
# reclassifying the SST raster for oyster suitability (11-30°C)
# first creating a reclassification matrix
rcl_sst <- matrix(c(-Inf, 11, NA,
11, 30, 1,
30, Inf, NA), ncol = 3, byrow = TRUE)
# then reclassify the matrix
sst_suitable <- classify(average_sst_degC, rcl = rcl_sst)
# reclassifying the depth raster for oyster suitability (-70-0)
# first creating a reclassification matrix
rcl_depth <- matrix(c(-Inf, -70, NA,
-70, 0, 1,
0, Inf, NA), ncol = 3, byrow = TRUE)
# then reclassify the matrix
depth_suitable <- classify(depth_newres, rcl = rcl_depth)
# finding locations where depth and sst are suitable for oysters
# making a raster multiplication function
mult_fun = function(rast1, rast2){
rast1 * rast2
}
# creating the raster that has NAs where both are not suitable, and 1s where depth and sst are suitable
depth_and_stt_suitable <- lapp(c(depth_suitable, sst_suitable), fun = mult_fun)Scaling to Locations of Interest (Ecological Economic Zones)
# data exploration - are these rasters of the same extent/crs etc? yes!
wc_regions <- wc_regions |> rename(Region = rgn)
depth_and_stt_suitableclass : SpatRaster
dimensions : 480, 408, 1 (nrow, ncol, nlyr)
resolution : 0.04166185, 0.04165702 (x, y)
extent : -131.9848, -114.9867, 29.99305, 49.98842 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source(s) : memory
name : lyr1
min value : 1
max value : 1 # first, make wc_regions into a raster
wc_regions_simp <- wc_regions |>
select("Region")
wc_regions_rast <- rasterize(x = wc_regions_simp, y = depth_and_stt_suitable, field = "Region")
# finding the suitable cells within each region
# first mask the region raster with the suitable cells (since not suitable cells are NA, our region raster will only have values where suitable)
wc_regions_mask <- mask(wc_regions_rast, depth_and_stt_suitable)
# lastly, displaying our suitable cells within each region that are suitable
tm_shape(wc_regions_mask) +
tm_raster()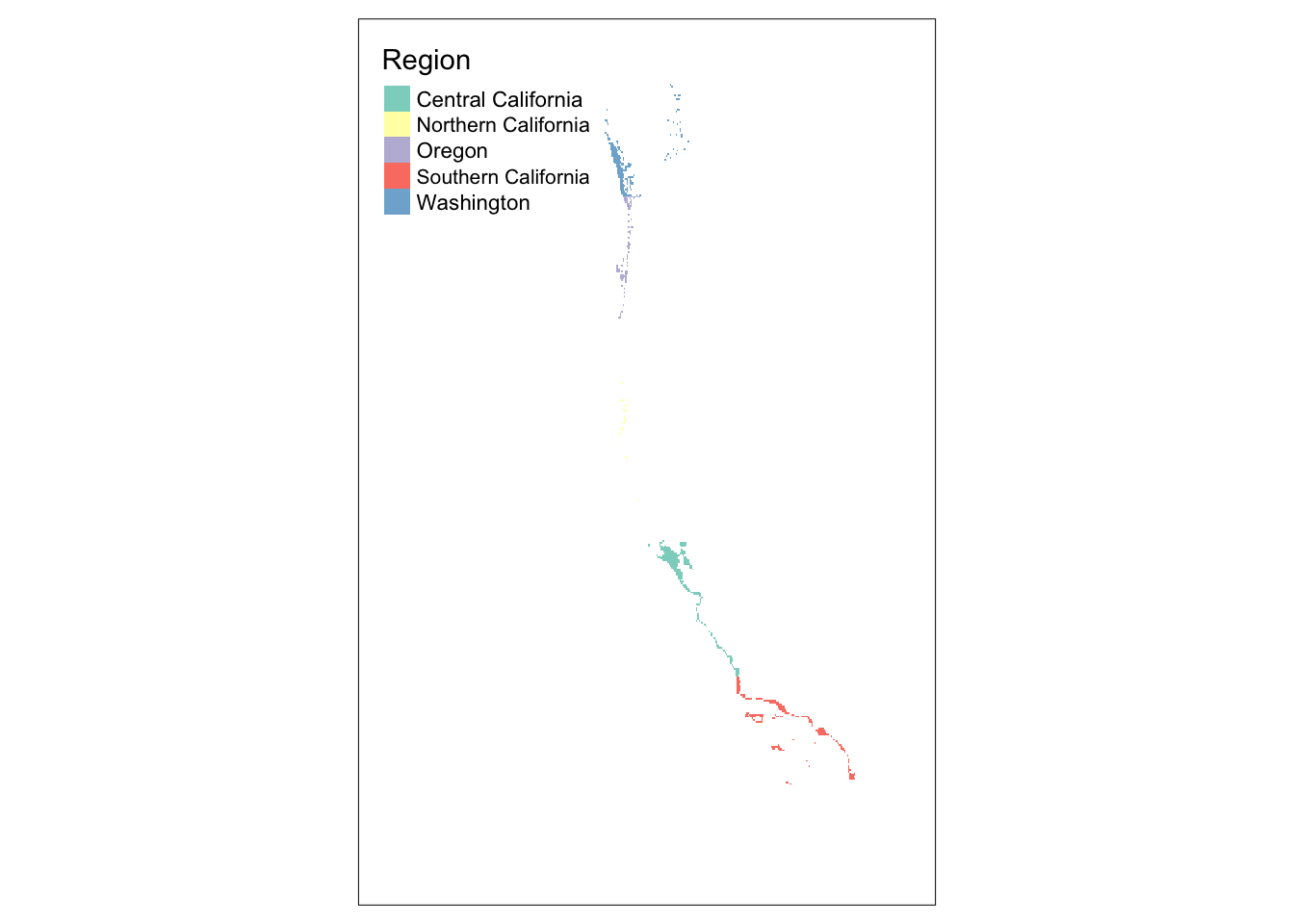
# finding the area of the suitable grid cells of each region
suitable_area_byregion <- zonal(cellSize(wc_regions_mask, unit="m"), wc_regions_mask, fun = "sum") |>
mutate(area = area / (1000^2)) |>
rename(suitable_area_km2 = area)
suitable_area_byregion Region suitable_area_km2
1 Central California 4923.1537
2 Northern California 438.1535
3 Oregon 1533.0949
4 Southern California 4096.5404
5 Washington 3224.7377# finding the area of the grid cells for each region (even though we have this already reported in the original datafile, we're using the same method we used to calculate our suitable area to make sure percentages are more likely proportional, since there are slight differences in this area approximation method).
total_area_byregion <- zonal(cellSize(wc_regions_rast, unit="m"), wc_regions_rast, fun = "sum") |>
mutate(area = area / (1000^2)) |>
rename(total_area_km2 = area)
total_area_byregion Region total_area_km2
1 Central California 202779.85
2 Northern California 163715.00
3 Oregon 179866.42
4 Southern California 206535.86
5 Washington 67813.69# find the percent of suitable area per area per region
percent_area_df <- inner_join(total_area_byregion, suitable_area_byregion, by = "Region") |> mutate(percent_suitable = (suitable_area_km2/total_area_km2) * 100)
# source code: https://stackoverflow.com/questions/68979884/how-to-loop-through-a-dataframe-and-get-value-of-corresponding-column
for(i in 1:dim(percent_area_df)[1]) {
writeLines((paste0(percent_area_df$Region[i], " has an area suitable for oysters of " , round(percent_area_df$suitable_area_km2[i], 2), "km2 which amounts to an area that is ", round(percent_area_df$percent_suitable[i],2), "% suitable for oysters.","\n")))
}Central California has an area suitable for oysters of 4923.15km2 which amounts to an area that is 2.43% suitable for oysters.
Northern California has an area suitable for oysters of 438.15km2 which amounts to an area that is 0.27% suitable for oysters.
Oregon has an area suitable for oysters of 1533.09km2 which amounts to an area that is 0.85% suitable for oysters.
Southern California has an area suitable for oysters of 4096.54km2 which amounts to an area that is 1.98% suitable for oysters.
Washington has an area suitable for oysters of 3224.74km2 which amounts to an area that is 4.76% suitable for oysters. # test: look at percentages using the given areas to see if theyre similar:
#percent_area_df_originalareas <- merge(wc_regions, suitable_area_byregion, by = "Region") |> mutate(percent_suitable = (suitable_area_km2/area_km2) * 100)Visualizing Suitable Ecological Economic Zones
# first joining the the area_df back to the region sf object for plotting
region_stats_join <- merge(wc_regions, percent_area_df)
# switching to tmap view mode to add a built in basemap
tmap_mode("view")
# total suitable area by region
# create legend title
legend_title_area = expression("Suitable Area for the <br> Oyster Species (km<sup>2</sup>)")
#legend_title_area = expression("Suitable Area for Oysters (km^2)")
area_suitable_map <- tm_shape(region_stats_join) +
tm_fill(col = "suitable_area_km2",
title = legend_title_area,
style = "pretty",
palette = "Blues") +
tm_borders() +
tm_legend(legend.outside = TRUE) +
#tm_layout(main.title = "West Coast Regions Suitable for Oysters",
#main.title.size = 1) +
tm_text("Region", size = 0.5) +
tm_basemap("CartoDB.Positron")
# percent suitable area by region
percent_suitable_map <- tm_shape(region_stats_join) +
tm_fill(col = "percent_suitable",
title = "Percent of Region Suitable <br> for the Oyster Species",
style = "pretty",
palette = "Blues") +
tm_borders() +
tm_legend(legend.outside = TRUE) +
# tm_layout(main.title = "Percent of West Coast Region Suitable for Oysters",
# main.title.size = 1) +
tm_text("Region", size = 0.5) +
tm_basemap("CartoDB.Positron")
tmap_arrange(area_suitable_map, percent_suitable_map)Creating a Function to Broaden this Workflow
This sort of analysis can be applicable for many different applications. Not only can this analysis be used to assess aquaculture suitability for other species, not just the oyster, but it can also be used to assess critical habitat needs for any key marine life that depends on certain sea surface temperature and ocean depth.
Here I turn my analysis into a function that takes in the marine species of interest, habitat temperature and depth ranges and returns maps of total suitable area and percent suitable area per exclusive economic zone.
Below I test my function using data from SeaLifeBase to find suitable habitat for the Giant Rock Scallop, usually found on the west coast of the USA.
# creating a function that takes in depth ranges, temperature ranges, and species name
suitability_map_fun = function(shallowest_depth, deepest_depth, min_temp, max_temp, species_name){
# code sourced from the "find suitable locations" section
rcl_sst <- matrix(c(-Inf, min_temp, NA,
min_temp, max_temp, 1,
max_temp, Inf, NA), ncol = 3, byrow = TRUE)
sst_suitable <- classify(average_sst_degC, rcl = rcl_sst)
rcl_depth <- matrix(c(-Inf, -deepest_depth, NA,
-deepest_depth, shallowest_depth, 1,
shallowest_depth, Inf, NA), ncol = 3, byrow = TRUE)
depth_suitable <- classify(depth_newres, rcl = rcl_depth)
mult_fun = function(rast1, rast2){
rast1 * rast2
}
depth_and_stt_suitable <- lapp(c(depth_suitable, sst_suitable), fun = mult_fun)
# code sourced from the "Determine the most suitable EEZ" section
wc_regions_simp <- wc_regions |>
select("Region")
wc_regions_rast <- rasterize(x = wc_regions_simp, y = depth_and_stt_suitable, field = "Region")
wc_regions_mask <- mask(wc_regions_rast, depth_and_stt_suitable)
suitable_area_byregion <- zonal(cellSize(wc_regions_mask, unit="m"), wc_regions_mask, fun = "sum") |>
mutate(area = area / (1000^2)) |>
rename(suitable_area_km2 = area)
total_area_byregion <- zonal(cellSize(wc_regions_rast, unit="m"), wc_regions_rast, fun = "sum") |>
mutate(area = area / (1000^2)) |>
rename(total_area_km2 = area)
percent_area_df <- inner_join(total_area_byregion, suitable_area_byregion, by = "Region") |> mutate(percent_suitable = (suitable_area_km2/total_area_km2) * 100)
# code sourced from the "Determine the most suitable EEZ" section
region_stats_join <- merge(wc_regions, percent_area_df)
tmap_mode("view")
legend_title_area = paste0("Suitable Area for the <br>", species_name, " Species (km<sup>2</sup>)")
area_suitable_map <- tm_shape(region_stats_join) +
tm_fill(col = "suitable_area_km2",
title = legend_title_area,
style = "pretty",
palette = "Blues") +
tm_borders() +
tm_legend(legend.outside = TRUE) +
tm_text("Region", size = 0.5) +
tm_basemap("CartoDB.Positron")
legend_title_percent = paste0("Percent of Region Suitable <br> for the ", species_name, " Species")
percent_suitable_map <- tm_shape(region_stats_join) +
tm_fill(col = "percent_suitable",
title = legend_title_percent,
style = "pretty",
palette = "Blues") +
tm_borders() +
tm_legend(legend.outside = TRUE) +
tm_text("Region", size = 0.5) +
tm_basemap("CartoDB.Positron")
tmap_arrange(area_suitable_map, percent_suitable_map)
}
# running the function for the USA giant rock-scallop
# source: https://www.sealifebase.ca/summary/Crassadoma-gigantea.html
suitability_map_fun(shallowest_depth = 0, deepest_depth = 80, min_temp = 0, max_temp = 22, species_name = "Giant Rock Scallop")References
Footnotes
Exclusive economic zones represent areas or “zones” of ocean that are assigned to a specific country for exclusive economic purposes such as fishing.↩︎
GEBCO Compilation Group (2022) GEBCO_2022 Grid (doi:10.5285/e0f0bb80-ab44-2739-e053-6c86abc0289c).↩︎
Citation
@online{cutler2023,
author = {Victoria Cutler},
editor = {},
title = {Finding {Suitable} {Locations} for {Marine} {Aquaculture}},
date = {2023-07-20},
url = {https://victoriacutler.github.io/posts/suitable-habitats/},
langid = {en}
}